
This blog charts the challenges, successes and discoveries of the Herts DNA Barcoding Hub at the University of Hertfordshire’s Bayfordbury Campus. The hub was initiated by Adam Hillier with support from the Darwin Tree of Life Enabling Communities Fund and Dr. Sam Rowe at the Earlham Institute. Adam will be writing regularly on the hubs progress and offering tips for others who want to establish a regional DNA barcoding hub with DToL.
If you would like to contact Adam about his experiences setting up a DNA Barcoding Hub, you can do so here.
Acknowledgement: Since the hub was opened in January activities we are very grateful to the University of Hertfordshire staff for welcoming Adam and members of the community to Bayfordbury and for their consideration for the team’s health and safety. Special thanks to Ronni Edmonds Brown, Bo Liu, Ian Flack, Clare Smith, Heather Fell and Aiden Bygrave. The hub is still in a very early stage but it is already attracting a good amount of curiosity, interest and ideas.
March 2023
The Herts Barcoding Hub assembles
In just two months, the work funded by the Darwin Tree of Life ‘Enabling Communities Fund’ and supported by DToL partners at the Earlham Institute has demonstrated that it is possible for Citizen Scientists to stimulate and lead local environmental DNA activities within a community.

‘Community pioneers’: Enthusiasts and experts
Working sessions so far have been restricted to individual or dual sessions with ‘community pioneers’. Larger workshops, with up to 8 people, are being planned for spring/summer once we have practised our multi-tasking skills!
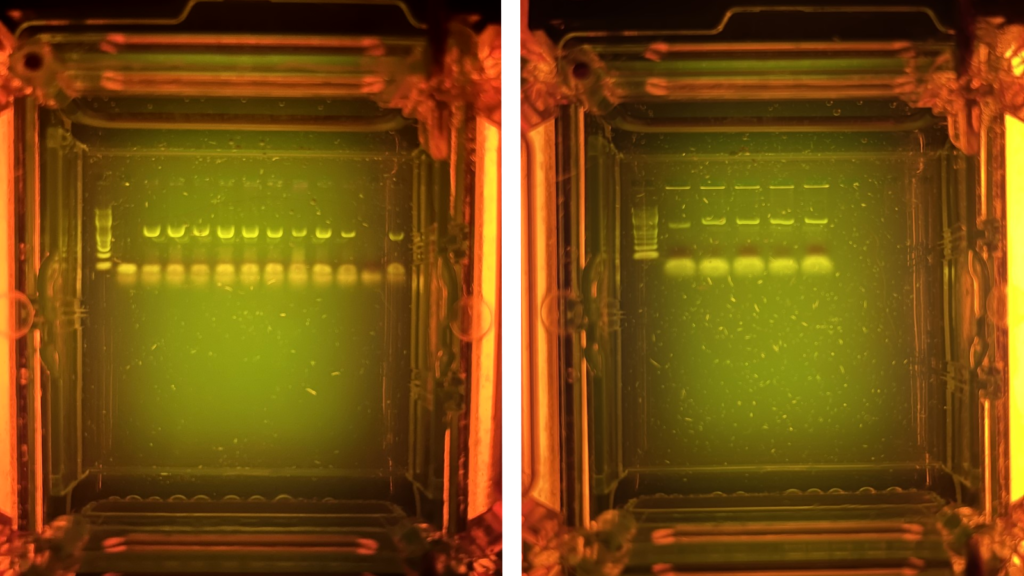
In total 10 sessions have been held so far looking at local fungi (5 sessions), plants (3 sessions) and insects (2 sessions). Whilst the fungi sessions have focused on real specimens of interest to local mycologists, the plant and insect sessions have been run to test the robustness of our workflows for different types of specimens. For example, we’ve tested different taxa (groups of related organisms), different states of preservation, and more.
 Fungi
Fungi
‘Success rates’ using the Bento Lab DNA extraction kit and focussing on the ITS barcode region are beginning to stabilise at around 70% (for extraction and amplification success) and 70% (for species level specimen ID success). Combining these two independent factors means that, on average, there is around 50% chance of generating a species level identification for a particular fungi specimen.
Some sequences can only be confidently identified to genus level, which can nevertheless still be useful. There have been no issues so far with contamination.
 Plants
Plants
The equally simple Chelex extraction method is being used along with a slightly modified protocol for waxy leaves. So far work has focussed on the rbcl barcode region. Ignoring some initial failures due to basic learning mistakes, plant DNA extraction and PCR amplification success rates are about 70%. However, outside positive controls, confident identification has only been possible to genus level.
Researchers from the Royal Botanic Garden Edinburgh are helping to provide guidance on which families of plants are likely to have the highest success rates. One option is to incorporate more barcode regions into the analysis but the plan is to focus on plant families/genera of local interest which have proven by research to have good success rates using rbcl and matk.
In the meantime I am using DNA barcodes sequences from selected native plants in Hertfordshire to learn about the evolution of flowering plants, which first appear as fossils in the Cretaceous Period.
 Insects
Insects
For insects, we are using the Chelex extraction method with PCR amplification focussing on the CO1 barcode region. Seven samples have been run but only one sample, a bee leg, resulted in a successful PCR amplification. Nevertheless, this one success demonstrates that the basic protocol and chemicals are all working.
Over the next few weeks, work will focus on workflow testing and optimisation using positive controls (aphids and ants) before moving on to more diverse and potentially challenging taxa.
In all cases successfully amplified DNA samples have been sent to the Azenta/Genewiz labs with sequencing results sent by email within 24 hours. With good quality sequences the bioinformatics step is very simple and can be done in a matter of minutes.

Building on success with new projects
Already plans are underway to scale up and diversify the hub activities. One example is with environmental DNA (eDNA) which monitors the presence of genetic material shed by various organisms into their environment, for example as skin, hair, faeces or mucous. A benefit of this is that sampling becomes non-destructive, simply taking a soil or water sample, for example, rather than the entire organism.
This work would focus on Bayfordbury ecological assessment and monitoring studies and be funded by the University of Hertfordshire. Demonstrations and research projects are being planned with Dr Bo Liu for the spring/summer which will likely pave the way for more comprehensive studies and applications in 2024 and beyond.
Additionally, Bayfordbury has been accepted as a BIOSCAN partner and work is planned to commence in April or May following a connection made at a Darwin Tree of Life event in January. As with other BIOSCAN sites, the work will involve collection of flying insects over a 24 hour period every month using Malaise traps, which our team will then sorted by size and taxonomic order. The BIOSCAN team at the Wellcome Sanger Institute apply large scale metabarcoding techniques to create species lists and, ultimately, develop ways to glean insights into insects’ interactions with their ecosystem.

Discussions are underway as to how the DNA hub facilities can be used for other side projects. But before we get carried away, let’s look at how DNA barcoding is progressing at the site.
January 2023
DToL’s DNA Barcoding inspires citizen science in Hertfordshire
Over the past two years, 183 budding genomicists have been trained in the scientific technique of DNA barcoding as part of Darwin Tree of Life’s public engagement activities. This took place across 25 workshops at the Earlham Institute in Norfolk, as part of their ‘Barcoding The Broads’ project.
DNA barcoding is an accessible and relatively low-cost science that allows anyone to easily and accurately identify species collected in the field by reading short sequences of their DNA – not dissimilar to the idea of scanning an item in a supermarket.

And now DNA barcoding techniques and technologies are being deployed and taught at a second location, the Bayfordbury Field Station at the University of Hertfordshire. Promisingly, Hertfordshire is already a county with a very active network of naturalists and life science research.

This new initiative is pioneering for Darwin Tree of Life in the sense that it is led locally by a citizen scientist, myself, with an earth science background but no prior experience in taxonomy or molecular biology – just the training provided at the Earlham Institute. If successful this will pave the way for democratising DNA barcoding across the UK through similar initiatives in other regions and nations.
The initiative in Hertfordshire is in the early stages but several solid foundations have already been laid in several key areas.
Building strong community relationships
The Bayfordbury DNA Hub has been set up following engagement with a variety of life science and active natural history groups within Hertfordshire. The initiative was launched in November with a 1 day workshop attended by representatives from the University of Hertfordshire’s Department of Life Science, the Rothamsted Research, the Hertfordshire and Middlesex Wildlife Trust, the Hertfordshire Natural History Society and the Heartwood Forest Long Term Monitoring Group. Within the initial group of interested experts there is a wide range of taxonomic and ecological interests in the community including birds, mammals, insects, plants, fungi and even meiofaunas such as nematodes.

Provide opportunities for learning / development
Being based at the University of Hertfordshire Bayfordbury Life Science Field centre will provide the opportunity for Life Science students to learn about DNA barcoding and to develop research project ideas. In addition, local naturalists will be encouraged to use the Hub for themselves after workshop-based training.

Application use cases
It is expected that much of the initial work will focus on the identification of specimens provided by local naturalists with priority given to rare/cryptic specimens. It is also expected to initiate a ‘Barcoding Bayfordbury’ project focussing on unique flora and fauna around the Bayfordbury site including the Clinton-Baker Pinetum. Other emerging ideas include identifying ‘purest native’ specimens of often hybridised plants (crab apples and brambles for example) for conservation purposes and leveraging the immense diversity of flowering plants in Hertfordshire to try to answer questions about the timing of evolutionary events.
It is expected that new ideas will be created in the first half of 2023 through workshops with local naturalists and Life Science university students.

 Fungi
Fungi Plants
Plants Insects
Insects