100 genomes annotated: EMBL-EBI reaches major milestone
Researchers at EMBL’s European Bioinformatics Institute (EMBL-EBI) are supporting Darwin Tree of Life by storing and annotating the genomes sequenced by the project, and making this data openly available through the DToL Data Portal. They have now hit a first big milestone in the project – putting together genome annotations for 100 new species.
Much like other types of annotation, genome annotation adds a useful explanation of what we’re looking at. In this case, researchers identify all the genes and coding regions in a genome sequence and try to determine what they do. Put simply, all our genome sequences need to be annotated before we can make sense of them.
We take a look at the challenges EMBL-EBI have overcome so far and their future plans within the project.
The DToL Data Portal
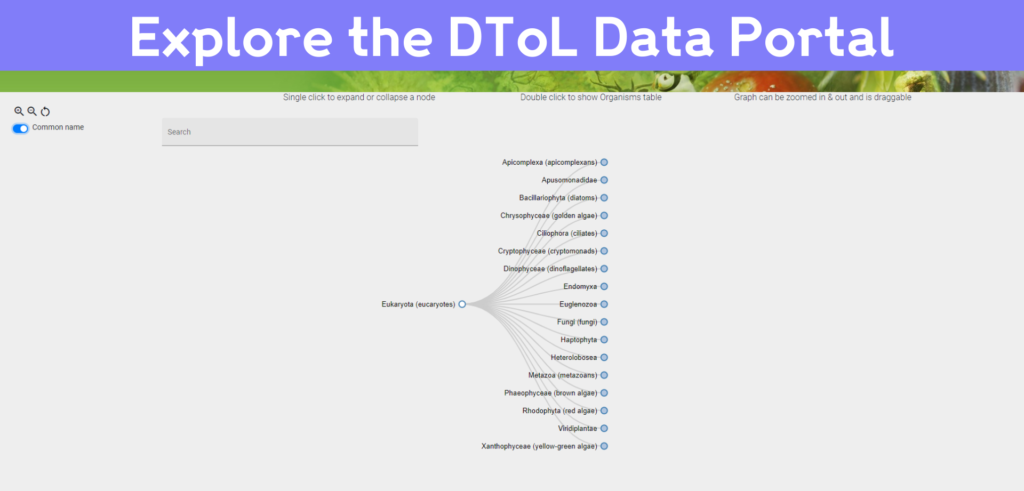
The DToL Data Portal serves both the scientific community and the public by showcasing the huge range of data generated by the project. It pulls together the sampling carried out by the different DToL project partners across the UK, all of the genome assemblies produced at the Wellcome Sanger Institute, and the annotation work conducted by the Ensembl team at EMBL-EBI.
The Data Portal itself was developed by Alexey Sokolov’s team at EMBL-EBI. As well as hosting all of the data from the DToL project and giving users open access to genome assemblies and annotations, this portal has a tracking feature where users can follow the sequencing progress of their species of interest. It also contains a phylogeny browser, using the evolutionary relationships between species to allow users to navigate the species in the portal along the branches of the tree of life.
“We’re constantly improving the Data Portal so the scientific community can get the most out of the Darwin Tree of Life data,” says Alexey Sokolov, Project Lead at EMBL-EBI. “The Portal currently allows users to track the status of their species of interest and we are working to make this process more detailed. Hopefully, in the future, we will include a sign-up for notifications about the status of particular species.”
Open access to the DToL data
The European Nucleotide Archive (ENA) team plays a vital role in making the DToL data open access and freely-available to the scientific community. They are also working to ensure the long-term storage of the DToL data in a standardised way.
To do this, the ENA is working to improve metadata standards for biodiversity data such as those generated through the DToL project. Metadata are a set of data that describes and gives information about other data. For example, DToL genome sequences will include mandatory spatio-temporal metadata (where and when a sample was collected) for all new data submissions. This also enriches the scientific value of the data for researchers worldwide.
“The ENA is adapting its data submission process to meet the needs of researchers working on global biodiversity projects such as Darwin Tree of Life,” said Josie Burgin, Bioinformatics Project Manager at EMBL-EBI. “Making these data open, findable, and reusable relies on having rich and relevant metadata.”
Genome annotations for biodiversity research
Rapid access to the genome annotations produced from the DToL project will have a huge impact on global biodiversity research by opening new doors for scientists in this field. Lepidoptera – butterflies and moths – and Hymenoptera – bees, wasps, and ants – are some of the first DToL genome annotations to be completed by the Ensembl team. Furthering our understanding of Hymenoptera genomes could help in the fight to prevent the devastating global decline of wild bee species.
“The annotation work that Ensembl does has scaled up massively to keep up with the data generated from the DToL project,” said Peter Harrison, Genome Analysis Team Leader at EMBL-EBI. “Our first big push was to get the genome annotations for Lepidoptera and Hymenoptera out to help researchers with their global conservation efforts. These were also all annotated in a matter of days which is an incredible turnaround compared to what we were able to do previously.”

The next big DToL challenge faced by the Ensembl team will be the arrival of several new plant species needing genome annotations. Plant genomes are often very different from animal genomes; their introns and genes are usually much smaller on average. This creates problems for Ensembl’s existing pipelines with optimised settings for an expected gene size. Some plant genomes are also gigantic – up to 40 times bigger than the human genome – making them tricky to work with and needing much more data storage.
“Things start to move more quickly once we have pipelines set up to run a particular group of species. It’s initially a very involved process and somebody has to test and check every step to make sure everything looks consistent,” said Fergal Martin, Eukaryotic Annotation Team Leader at EMBL-EBI. “Now we are at a stage where our pipelines will produce good genome annotations in an extremely short timeframe for the species we have cracked. For example Lepidoptera and Hymenoptera; we’ve put together a lot of genome annotations for these species and so it’s much more straightforward to create new genome annotations when more bees or butterflies start to come our way.”
With hundreds more genome assemblies set to emerge from the DToL pipeline in 2022 alone, there will be plenty of work for the EMBL-EBI team to get stuck into over the coming months and years of the project.
Victoria Hatch is Communications Officer at EMBL-EBI. This article was adapted from an original version on the EMBL-EBI website.