Life inside life: Understanding the ‘pandemic’ infecting nearly half of all insect species
When Darwin Tree of Life’s entomologists – the insect experts – venture out into the woods and meadows of Britain and Ireland, they may only be seeking a certain species of butterfly or beetle. But that’s not all that gets swept up in their nets.
Living on and within these ‘target’ species are a plethora of symbiotic and parasitic microbes, bacteria and viruses, collectively known as ‘cobionts’. These too end up snap-frozen and sent to the lab, destined for our genome-sequencing processes.
DToL is ostensibly only targeting eukaryotic organisms: those whose cells have a nucleus, including all animals, plants, fungi and protists – but excluding bacteria and viruses.

However, with hundreds of different insect specimens from across Britain and Ireland arriving at their lab, researchers at the Wellcome Sanger Institute’s Tree of Life Programme could not resist the opportunity to study their bacterial cobionts as well – getting two (or more) genomes for the price of one.
Of particular interest was Wolbachia, a bacterial ‘pandemic’ which is thought to infect nearly half of all insect species. A new Wolbachia study published in PLOS Biology, looking at 368 insect species collected for DToL, has provided a broader understanding of how this ubiquitous bacteria evolves alongside and interacts with its insect hosts.
A complex pandemic
First things first: when thinking about the Wolbachia pandemic in insects, try not to draw too many parallels with the pandemics that affect our own species. This bacteria is not causing mass insect mortality, although it can significantly impact insects’ biology.
For the Wolbachia, the relationship is straightforward. There are no known free-living Wolbachia strains, and without a host they cannot survive. For the insect, things are more complex. In many species, infection by Wolbachia can alter their biology but still be beneficial. In others it is inconsequential.
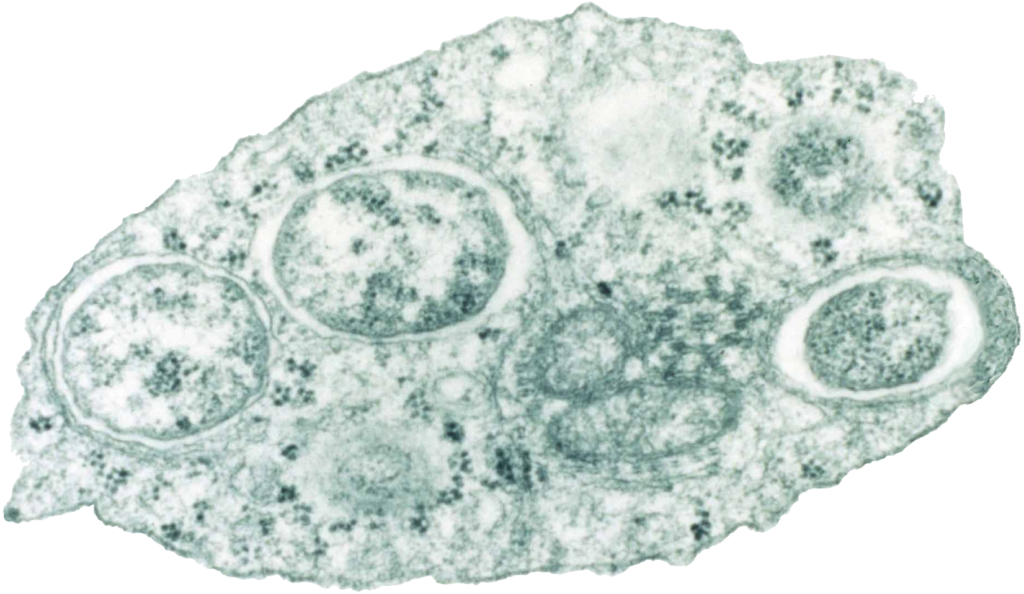
Take mosquitos as an example. Infection by Wolbachia can trigger a phenomenon called cytoplasmic incompatibility, which alters the mosquitos’ reproductive capabilities. Infected females can mate with all males, but uninfected females can only mate with uninfected males. While this is not an existential problem for the mosquito species, it does help the Wolbachia to spread. Meanwhile, Wolbachia can also boost immunity against other microbial infections, including from the Plasmodium parasite that causes malaria. This is beneficial to the mosquito.
Another benefit in different groups of insects is how some Wolbachia strains can produce biotin, more commonly known as vitamin B7. This seems to be particularly beneficial to insects with nutrient-deficient diets, such as those that feed exclusively on nectar.
Wolbachia doesn’t just infect insects, it also affects nematodes. Elimination of their Wolbachia cobionts turns the nematodes sterile, making them unable to reproduce. Antibiotics targeting Wolbachia are therefore an effective treatment for parasitic nematodes that infect humans.
The complex effects of Wolbachia infection have a clear unifying feature: they all boost the bacteria’s selfish, Darwinian desire to spread. But it has been so successful, across so many species, mutating into so many different strains, that broader studies of its impacts have proven difficult – until the Darwin Tree of Life project offered a perfect opportunity.
Infection across the tree of life
Wolbachia has been studied for decades. But the Darwin Tree of Life project has allowed this latest research, by Dr. Emmelien Vancaester working within the Blaxter Group at Sanger’s Tree of Life Programme, to explore an unprecedented breadth of Wolbachia-infected species.
Ultimately, DToL aims to produce high-quality reference genomes for tens of thousands of insect species across Britain and Ireland. Emmelien, however, had to make do with the first 368 species to be sequenced – still a large and diverse sample.
In total, Emmelien managed to produce 110 complete Wolbachia genomes from 93 of the hosts. The host species belonged to 92 different insect families across 7 insect orders.
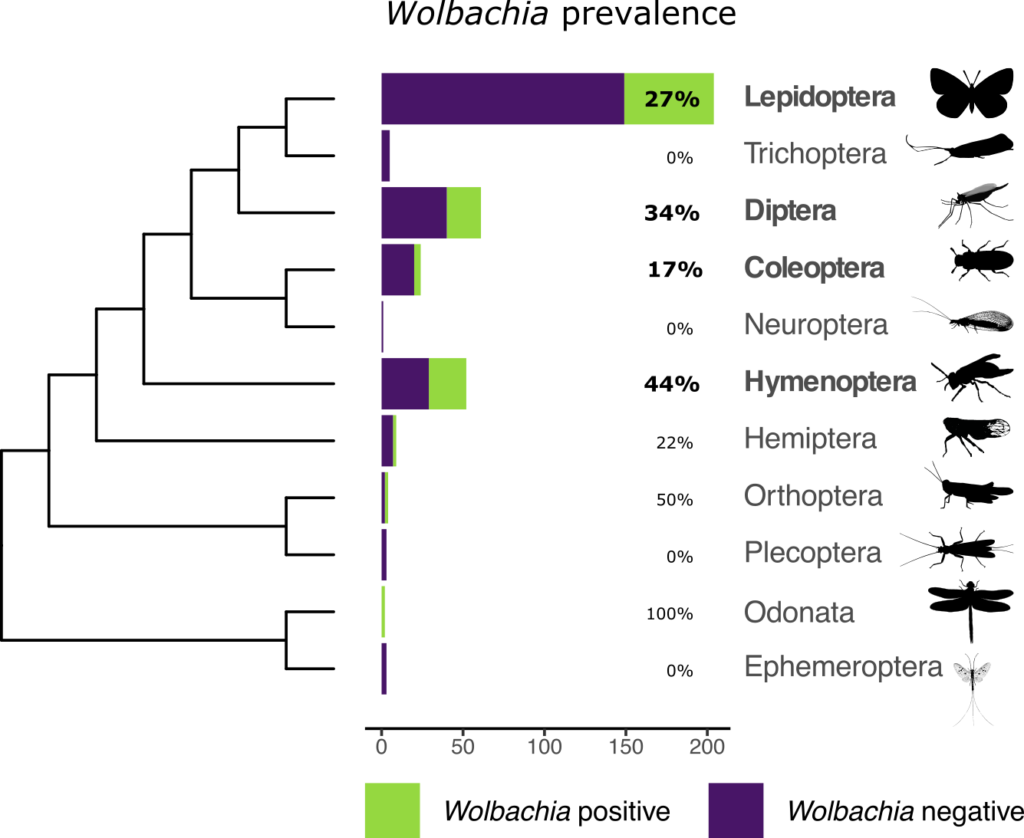
This study has more than doubled the number of insect host species for which Wolbachia genomes are available. It also provides the first Wolbachia genomes to be studied from the orders Odonata (dragonflies and damselflies) and Orthoptera (grasshoppers, locusts and crickets).
The Wolbachia genomes are not just a great collection of new data: by looking more deeply into the sequences and their relationships, Emmelien made some interesting discoveries.
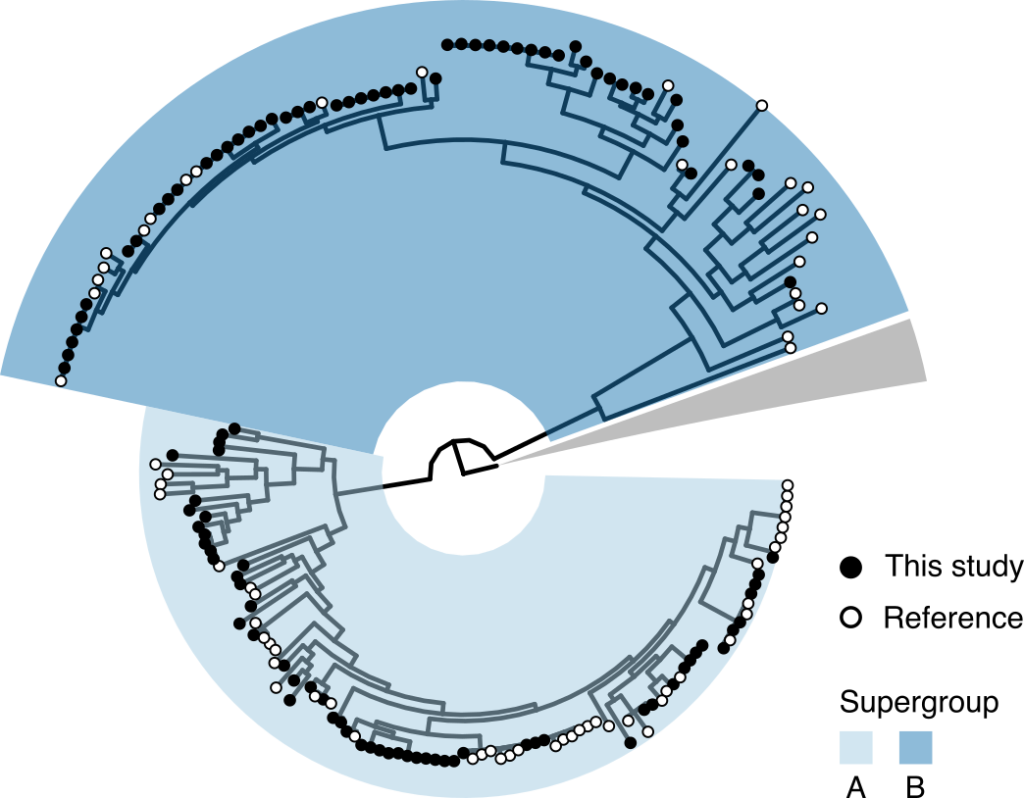
The link between Wolbachia strains and insect species is a bit of a mess. The study showed a distinct lack of what evolutionary biologists refer to as ‘cophylogeny’, where two species evolve closely together. Essentially, across evolutionary time the Wolbachia have not been faithful to their insect hosts, with many incidences of ‘host-switching’ by Wolbachia strains. Sometimes this host-switching wasn’t even within the same order of insects – for example, it might have switched from a fly to a moth.
The aptly-named tanglegram below shows how closely related insects (on the left) are not necessarily infected by closely related Wolbachia. The Wolbachia strains are categorised into eight ‘supergroups’, of which supergroups A and B are most common in insects. The study showed that, while strains from one supergroup tend to be more common in a certain insect order, this is by no means a hard-and-fast rule.
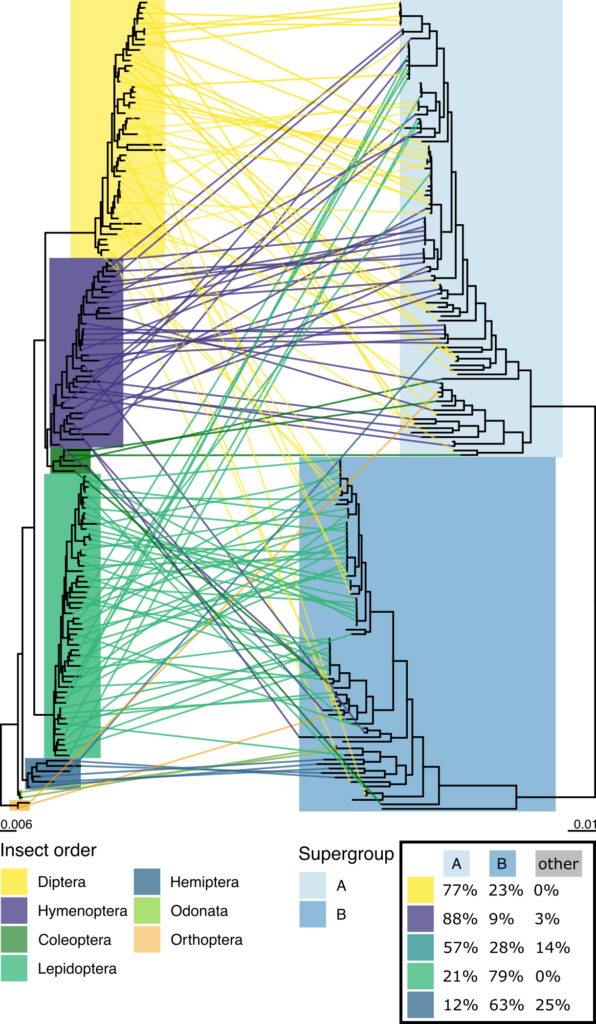
Host-switching may be driven by insect ecology. A tantalising clue to the pattern of host-Wolbachia association is that, of the seven insect species infected by the biotin-producing Wolbachia, three were from the genus of bees Andrena and another was from the genus Nomada. Nomada is a kleptoparasite, laying its eggs in the nests of Andrena species and stealing the food its unwitting hosts have already collected. It is possible the closeness of this ‘cuckoo’ behaviour has allowed the Wolbachia to switch species hosts.
Geographical location seems to have little impact on the pattern of Wolbachia infection. Increasingly, DToL will be collecting insect species from all over Britain and Ireland. But at this early stage in the project 82% of the 368 species studied were collected thanks to the tireless efforts of the DToL team in Wytham Woods near Oxford. There was no sign that samples collected at Wytham carried Wolbachia that were more closely related to each other than they were to Wolbachia from hosts collected elsewhere. Wolbachia must spread efficiently across the landscape as it jumps from host to host.
The Wolbachia family tree has grown. An important aspect of the DToL project is its use of the latest PacBio long-read DNA sequencing technology to help generate the most accurate genome assemblies yet, with 77 of the Wolbachia analysed in this study producing fully complete, circular genome assemblies. New technology now allows researchers to independently assemble closely related Wolbachia strains with confidence.
Wolbachia genomes are slightly larger than we thought. It was discovered that Wolbachia genomes are 10% larger on average than we thought. Much of that extra size was hidden in repeated sequences which were harder to spot with other technologies.
An added bonus was the largest Wolbachia genome yet to be assembled – analysed from a Blue-tailed Damselfly (Ischnura elegans) and measuring 2.19 million bases in length.

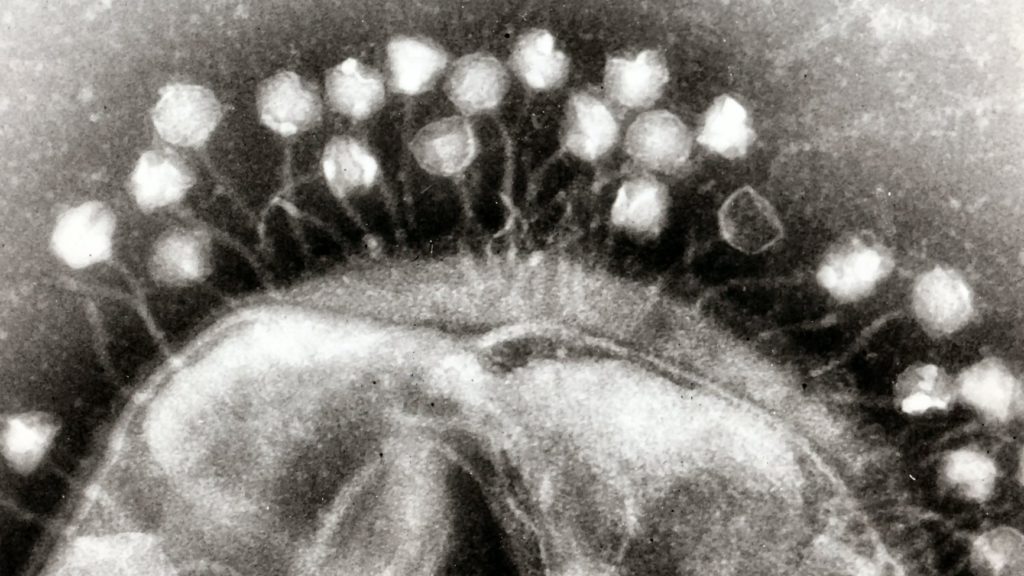
Wolbachia genome size is linked to their own tiny cobionts. Like a matryoshka doll of biodiversity, the Wolbachia bacteria living inside insects are themselves host to an even smaller virus. The presence of this bacteriophage was found to correlate with the size of the Wolbachia genome. The virus carries genetic material important to its own reproduction, which may be what is increasing the gene content of the host Wolbachia.
DToL’s microbial future
This study is not the end of DToL’s relationship with Wolbachia. Each insect that passes through the labs will have its Wolbachia analysed, and the dataset created in Emmelien’s research will continue to grow. The DToL team also looks for other microbial cobionts. While these are not encountered as frequently as Wolbachia, the scale of DToL means that they too will be accessible to comprehensive analysis.
As the project matures, there is also scope to answer new questions. How do habitat types impact Wolbachia infection? How do Wolbachia and other bacteria, such as Rickettsia or Spiroplasma, interact? And of course DToL is affiliated with a network of worldwide projects generating reference genomes for all life on earth, the Earth BioGenome Project. These kinds of studies have the potential to go global, and the tools developed for this work in DToL can be applied round the world.
Perhaps the most important thing this study has shown is that a massive biodiversity genomics project like Darwin Tree of Life offers unparalleled opportunities to study the microbiomes of all sorts of larger species.
“Biodiversity projects tend to talk about studying and comparing individual species, perhaps because that’s what people are more familiar with,” Emmelien says. “But I think this perception needs to shift, so we stop treating organisms as isolated objects and start seeing them as a network of life cohabiting in the same spaces and interacting across time.”
Thanks to Dr. Emmelien Vancaester and Prof. Mark Blaxter for their contributions to this article. The full paper, Phylogenomic analysis of Wolbachia genomes from the Darwin Tree of Life biodiversity genomics project, is published open access at PLOS Biology.