Sequencing the earthworms of Wytham Woods
In 2020, Liam Crowley contacted me from the University of Oxford about sequencing earthworms in Britain. Liam’s work on the Darwin Tree of Life project focuses on invertebrates at Wytham Woods, a 1,000-acre semi-natural woodland owned and maintained by the University of Oxford. Currently, he is working with others on the first phase of the project – to sequence the full genome of 2,000 species from as many different taxonomic families as possible. They are also focusing in greater depth on certain groups of particular ecological or evolutionary interest. Later phases will aim to ramp up this sequencing to eventually sequence every species in Britain and Ireland!

Recording earthworm genetic material
In the British Isles we have just 31 species of earthworm that occur in natural environments. This makes earthworms an easy target group for getting a good head start during phase 1 of the project. Twenty-nine of those species belong to a single family, Lumbricidae, with the remaining two species being the only species within their respective families in the UK (Acanthodrilidae and Sparganophilidae) and are both very rare and difficult to find — I’ve never personally come across either.
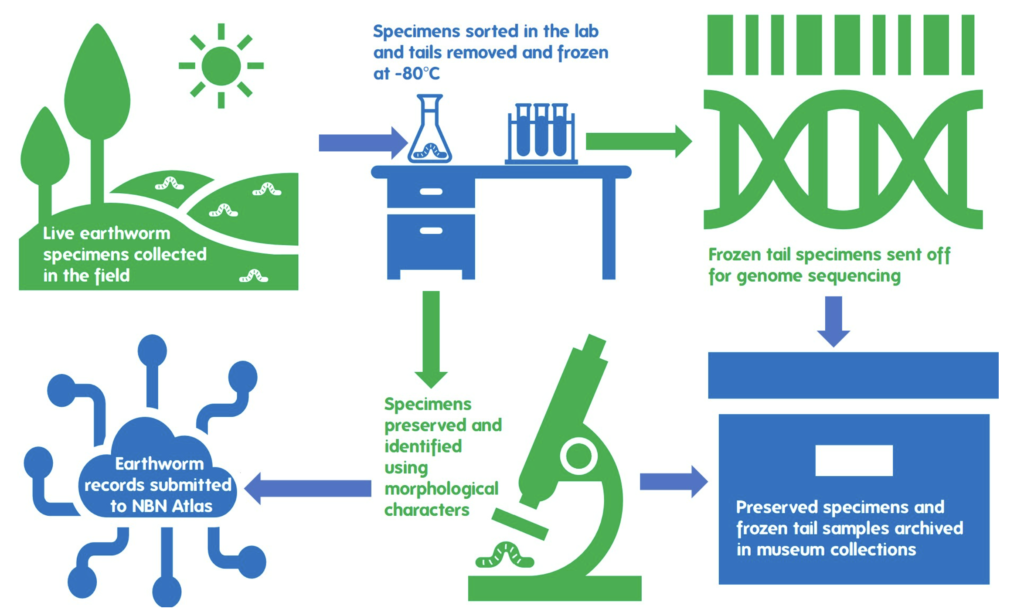
As live specimens are required to obtain the genetic material , my task was to find as many different species as I could over a two-day period while working on the project with Liam at Wytham Woods. I collected live specimens from my garden early in the morning of May 27, before heading to the Woods to undertake further sampling throughout that afternoon and next morning. On the second day we were joined by Michael Tansley, an Oxford PhD student studying earthworms.
In addition to my garden, we explored ancient woodland, calcareous grassland (at Wytham this derives from limestone) and fen wetland habitats, looking within soil, in and under deadwood and in the leaf litter layer. Identifying live earthworms in the field is extremely difficult and rarely even possible, so it was hard to know how many species we had collected. Any juveniles (earthworms with no clitellum or “saddle”) were released as it is not possible to identify their species — though this may be possible in the future using DNA barcoding.
Extracting genetic material from earthworms
The DToL project is using an exciting and relatively new sequencing technology known as ‘long-read’ sequencing, which reads and works out the order of nucleotides in much longer fragments of DNA than other methods. In much the same way as a jigsaw with fewer, larger pieces is easier to assemble than one with many small pieces, longer DNA fragments allow for a more accurate genome assembly.
The catch, however, is that DNA is a very unstable molecule, and starts breaking down into smaller fragments very quickly in dead tissue. To allow successful long-read sequencing, therefore, living tissue needs to be flash frozen to preserve long chunks of DNA. We achieved this for our earthworm samples by removing and immediately flash freezing a small section of the tail from each specimen at -80oC, before the specimens were euthanised and preserved in 80% ethanol.
A further small piece of the tail was preserved in 70% ethanol, to be submitted to the Natural History Museum, London, for DNA barcoding. The purpose of also barcoding specimens is three-fold: it allows us to populate the species barcode reference databases; allows matching of barcode ID against the identification made by collectors (preventing unnecessary expensive sequencing of the same species multiple times); and finally, allows a sense check that samples have not got mixed up during the sequencing process and each genome is matched to the correct specimen.

Once the relevant material was preserved for each molecular method, the remainder of the specimens (everything but a small piece of the tail) were identified under a stereomicroscope. I identified the specimen using the Key to the Earthworms of the UK & Ireland (2nd Edition) by Emma Sherlock. This key uses external morphological features such as the type of head, location of the male pore (reproductive organs), location and shape of the Tubercula Pubertatis or TP (a glandular thickening of the clitellum, thought to be used in mating), and the spatial distance between the setae (or “bristles”).
Ideally, we’d like to sequence every specimen to build up a more complete library of earthworm genomes. However, there is a cost to sequencing each sample, so we prioritised up to three specimens per species for sequencing. Those specimens that will not be sequenced were still identified and contribute important records to the National Earthworm Recording Scheme.

(Image: Liam Crowley, University of Oxford)
Survey outputs
The outcomes for our survey were as follows:
Earthworm species records
All specimens were identified and records submitted to the National Earthworm Recording Scheme. In total this contributed 43 species records across 14 species of earthworm, including 13 new species records for Wytham Woods, a previously unrecorded site. These records will be publicly available through the National Earthworm Recording Scheme (UK) dataset on the NBN Atlas.
Genome sequencing
Thirty-four specimens were sent for genome sequencing across 14 different earthworm species (see list below). This will enable different DNA extraction methodology to be tried and tested. This is necessary because earthworm biochemistry is quite different to other groups such as insects, for which we have the most knowledge of suitable methods, therefore different variations on extraction methods need to be tested. Any successfully sequenced genomes will be published with open access, making all the data publicly available for everyone.
- Allolobophora chlorotica
- Aporrectodea caliginosa
- Aporrectodea rosea
- Bimastos eiseni
- Bimastos rubidus
- Eisenia fetida
- Eiseniella tetraedra
- Lumbricus castaneus
- Lumbricus rubellus
- Lumbricus terrestris
- Murchieona muldali
- Octolasion cyaneum
- Octolasion lacteum
- Satchellius mammalis
Preserved specimens
Thirty-four preserved specimens were submitted to the Natural History Museum, London, earthworm collection, where they will be curated to a high standard and available for further inspection and research.
This article was originally published on Keiron Derek Brown’s website Biological Recording. Liam Crowley also contributed to and reviewed the article.
Keiron Derek Brown is an earthworm expert, chair of the Ecology & Entomology section of the London Natural History Society (LNHS) and project manager for the Field Studies Council’s BioLinks project.