From pondwater to single cells: 5 tricks for finding protists in the lab
This article is the second in a series exploring how the tiniest organisms Darwin Tree of Life is studying – the single-celled protists – are collected, processed and ultimately have their genomes sequenced. The first article focuses on collection in the field, Priest Pot: A seldom-studied protist paradise and the final article looks at How bioinformatics can crack the complex case of protist biodiversity.
Once our Darwin Tree of Life protist-hunters have returned from the field with litres of environmental water (e.g. pondwater) – as the University of Oxford team did at Priest Pot – the next task is to separate out the single-celled organisms we wish to study from the millions of diverse cells present in the water sample.
There are three types of protists – phototrophs, heterotrophs and mixotrophs.
Phototrophs are protists that make their own food and produce part of the oxygen we breathe by using the energy from sunlight and carbon dioxide (CO2). They are responsible for the giant colourful “blooms” we sometimes see on the surface of water.
Because they are so visible, grow in such large numbers, and are easy to maintain in laboratory cultures, phototrophs are the best-studied protists.
On the other hand, there are the heterotrophs, which move around and feed on other microorganisms, including predators and parasites of other protists and bacteria. These are much harder to spot, less studied as a result, and therefore highly sought after by our scientists.
Mixotrophs not only use energy from the sun to generate their own food and grow but they can also feed on the other types of protists. How mixotrophs feed often depends on the environmental conditions and sometimes both behaviours happen at the same time.

Ultimately, the team needs to separate and place the protists they want to study onto sample plates, each containing 384 tiny wells which will hold one single-celled organism. This method is known as ‘cell sorting’. Afterwards, the individual wells are imaged using a microscope and then these plates will be sent to the Earlham Institute, in Norwich, for genome and transcriptome sequencing.
But how do our researchers reach that point, having started with litres of water containing many cells belonging to all sorts of protists and other life?
1. Concentrating the water sample
In order to speed up the process of cell sorting, the pre-filtered water sample is first concentrated. Scientists select cells from 3μm up to about 50μm in length (about a 20th of a millimeter) using a technique called gravity filtration. This has a further advantage that the team can separate much of the smaller bacteria from the protists in their samples.
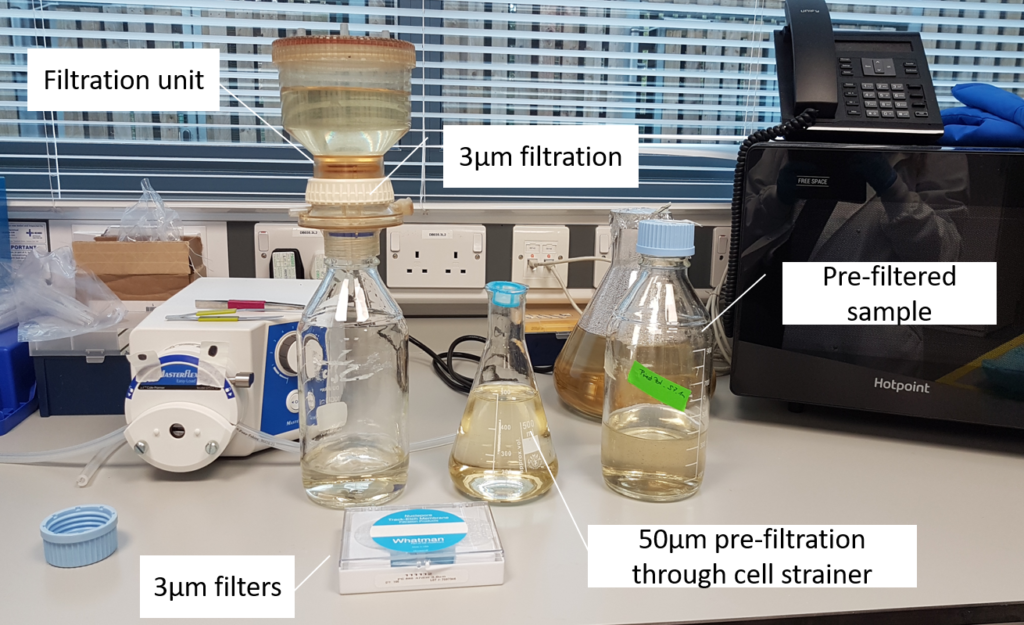
(Image: Estelle Kilias, University of Oxford)
- The sample first gets filtered through a 50µm cell strainer to remove larger particles that could cause clogging during cell sorting.
- Afterwards, the sample is concentrated by gravity filtration onto a 3µm filter.
- The target cells remain in the upper container while smaller particles and freshwater pass through the filter.
2. Detecting fluorescence
One way to identify and sort phototrophs is to detect their natural fluorescence due to the different pigments they contain. For this, we use a flow cytometry-based technique called fluorescence activated cell sorting (FACS). However, this technique alone is not helpful for spotting the less well-studied heterotrophs, which are not pigmented, and therefore non-fluorescent.

- The concentrated sample gets ‘cell sorted’ into a 384-well plate with one cell in each well.
- The cell sorter, “Melody”, sorts cells based on their fluorescence signal.
- Protists that are capable of doing photosynthesis (phototrophs and mixotrophs) have photopigments such as chlorophyll a. The cell sorter detects this signal and sorts the cells into the plate.
3. Using stains
Heterotrophs can only be spotted and told apart from debris by staining, with different stains used to identify different parts of the protists’ cell. Some examples include:
Nucleus staining – which helps differentiate protists from inorganic matter like soil, but again doesn’t split the phototrophs from the heterotrophs.
Flagellum staining – heterotrophs and mixotrophs use tail-like appendages to move around and hunt their prey. This stain separates flagellated from non-flagellated cells, but sometimes the signal is too weak.
Lysosome staining – this specifically targets the food vacuoles of heterotrophs or mixotrophs, which are used to digest their prey and are therefore acidic. The team are finding this method the most successful in their heterotroph/mixotroph hunt.
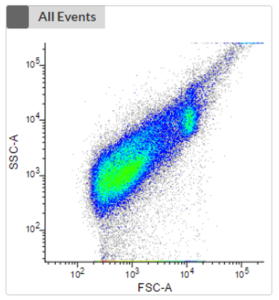
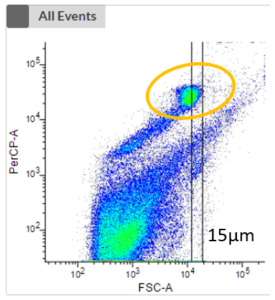
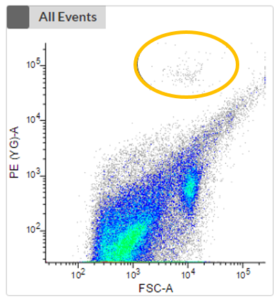
- Non-phototrophic protists (heterotrophs) require additional staining to distinguish cells from debris.
- There are different stains available, e.g. to stain either the nucleus or the food vacuole.
- The first plot shows an unstained sample without the consideration of any fluorescence signal of phototrophic cells. No clustering is visible, and it is impossible to distinguish phototrophic, mixotrophic or heterotrophic cells from each other or from debris.
- The next two plots take the fluorescence signal of two different photopigments into account. We can now see a clustering (highlighted in orange) which represents phototrophic cells that can now be cell sorted.
4. Darkness
Other tricks include sealing off the sample from light. This disadvantages the more abundant phototrophs which would otherwise dominate the sample. In turn, this benefits heterotroph and mixotroph species, which do not require light to grow and changes the diversity and population ratios within the sample.
5. Feeding
The specific prey of certain heterotrophs can also be encouraged to grow in order to increase the abundance of heterotrophs. The Oxford team has used a barley grain – first sterilised in an autoclave – to help increase the number of bacteria upon which certain heterotrophs like to feed. This increases heterotroph abundance and therefore helps their sorting.

- Each cell-sorted plate is imaged using a high content screener to assess the actual success of the cell sort.
- This involves a fluorescence image of each well being taken at a certain magnification.
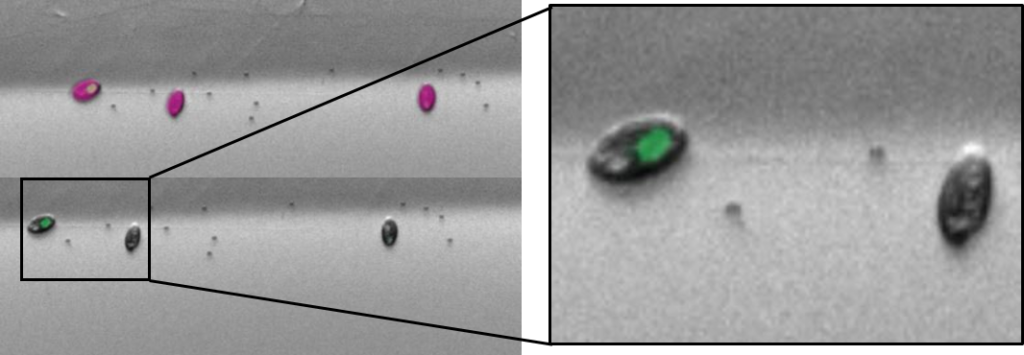
- To the left, you have an example of a multicell sort (multiple cells per well) of cryptophytes.
- Cryptophytes are an important food source for grazers due to their high nutrient value and small cell size. They can be found in freshwater, estuarine and marine ecosystems and are one of the taxonomic target groups in the Darwin Tree of Life protists working group.
Next steps for the Priest Pot samples
At time of writing, the DToL team at Oxford have sorted ten 384-well plates – each well containing one single-celled organism. After imaging, a cell lysis fluid is added to each well and the cell sorted plates are stored at -80C until further processing.
This processing will occur at the Earlham Institute, and the Oxford team expect to send the Priest Pot protists to Norwich in the next few weeks.
This is when yet more ground-breaking work for the Darwin Tree of Life project begins. Protists have invented a lot of different types of cell wall to protect them from their environments. Our scientists will need to find a way to crack them open to reach the genetic information – and different techniques will be required for different groups of protists.