How bioinformatics can crack the complex case of protist biodiversity
This article is the third in a series exploring how the tiniest organisms Darwin Tree of Life is studying – the single-celled protists – are collected, processed and ultimately have their genomes sequenced. The first article focuses on the collection of protists from Priest Pot, a pond in Cumbria, and the second looks at techniques to separate single cells from litres of pondwater.
Life is everywhere you look, it’s just very difficult to see most of it with the naked eye. Particularly fascinating are the strange and ubiquitous protists, single-celled eukaryotes that exist in more shapes and sizes than you’ve likely ever imagined.
Researchers at Earlham Institute and the University of Oxford are collaborating as part of the Darwin Tree of Life project to find, isolate and identify all of the protists in Britain and Ireland.
It’s a tricky task. Unlike with plants and animals, which tend to be readily observable and well documented already, we’ve got absolutely no idea how many protist species there are at all. The ones we do know about are complex enough as it is.
Thankfully, the Earlham Institute’s Dr Jamie McGowan and Jim Lipscombe are on the case.

Sequencing protist DNA is not easy
“We knew from the start there wasn’t going to be a one size fits all protocol,” says Lipscombe. “There are so many different kinds of cells. Diatoms are basically glass and tend to be particularly difficult to lyse open, while others seem to lyse open very easily – and all things in between.
“It’s a challenging project.”
Lipscombe’s main role is to prepare protist cell lysates (the fluid remaining after bursting cells open) for sequencing of their DNA and RNA, so that we can get an idea of both the genome and the transcriptome. He uses a modified version of G&T-seq, a protocol first developed by Dr Iain Macaulay at the Earlham Institute and widely used by the Single-Cell Genomics group.
Aside from the complexities of breaking the cells apart to access their genetic material, there’s also the precarious nature of working with single cells from environmental samples.
“You only have one shot for any given cell,” says Lipscombe. “When you’re sequencing from a pond, there might be hundreds of thousands of cells in a millilitre of water. But there might only be one or two of a particular kind that are of interest.
“The numbers are against us all the time, but it’s part of the fun.”
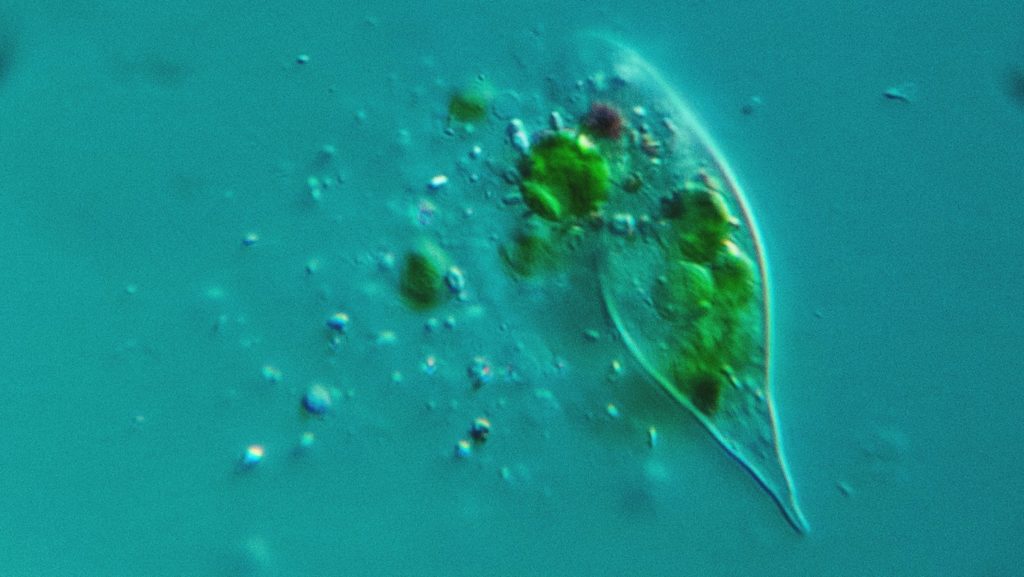
Assembling protist genomes using bioinformatics
Once the DNA and RNA have gone through the sequencers, it’s over to Dr Jamie McGowan in the Swarbreck Group. He spent his PhD studying protist parasites of plants, and has now turned his hand to decoding the lot of them.
McGowan’s role is to “analyse the sequencing data, assemble the protist genomes, work out how they’re evolving and how they’re related to each other, and then to build a picture of what they’re doing in different environments – what they’re interacting with.”
As with the sequencing, the bioinformatics portion of the research poses new challenges.
“Protists are massively understudied, so there’s very little genomic data available” explains McGowan. “A lot of people assume that, because they’re microbes, they might have really simple genomes.
“But some of these protists have genomes that are absolutely massive, or their genes are structured in really weird ways. Lots of the existing bioinformatics tools don’t really work well on them.”
Occasionally, protists can have larger genomes than our own. There are species of amoeba, for example, that purportedly have among the largest genome sequences in the world. That makes them challenging to both assemble and annotate.
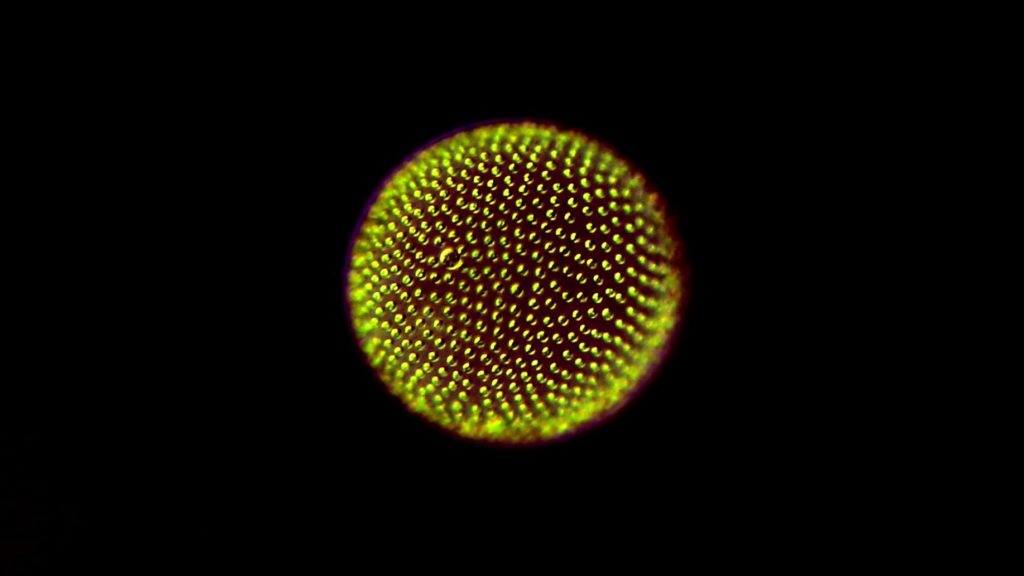
Discovering new genes and interactions with single-cell genomics
These challenges are what led McGowan and the team to tailor and apply techniques borrowed for other aspects of biology in order to paint a full picture of protist life.
“We’re taking single-cell genomics techniques that are typically used for biomedical research and applying them to study microbial biodiversity,” says McGowan. “Thanks to that, we can detect things like bacteria that the protists might have eaten, or viruses that may be infecting them.
“It’s a great application of single-cell genomics. These are things that, if we did traditional sequencing, we might not be able to detect because sometimes they’re present at really low abundance. With single-cell genomics, we can detect these interactions.”
McGowan is particularly interested in how the protists he’s sampling are related and how they’ve evolved to be the way they are.
“Once we have the genomes we can get an idea of what their metabolic potential is,” he explains. “That can tell us what they might be doing in a particular environment, why they are there. For many of the genes we find, they’re completely novel – so we have no idea what they do.
“With almost every single species we’ve sequenced so far, there’s been something unusual. It’s really cool and exciting.”
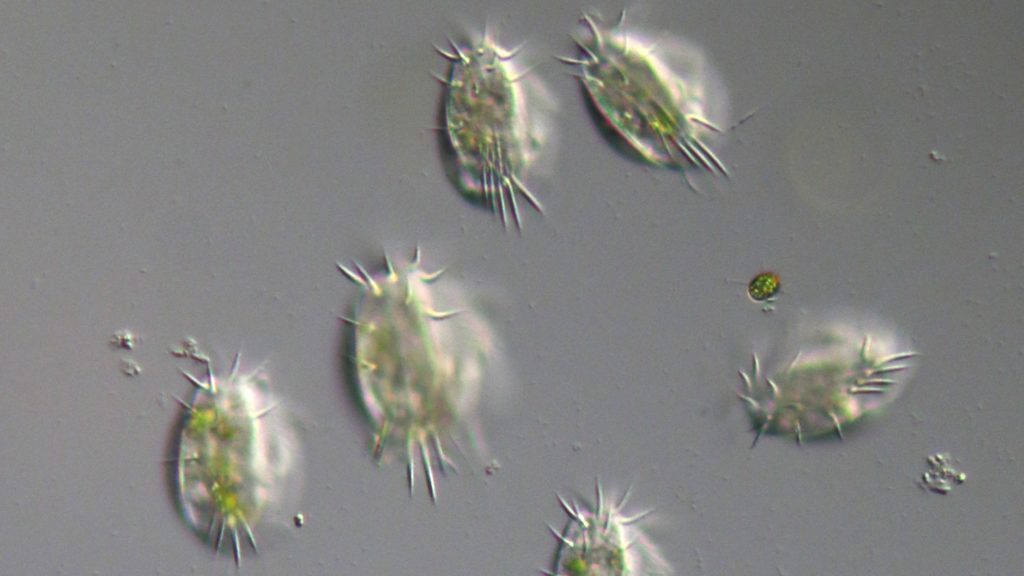
Scaling up to the whole protist tree of life
One pond is tricky enough at the moment, but the grand aim of the Darwin Tree of Life project is to sequence and analyse the DNA of every single eukaryotic species in the Britain and Ireland. Just how will we do that?
“We’re trying to scale everything up,” says McGowan. “Everything will be automated in the future. Jim is automating the labwork, while I’m automating the bioinformatics tasks. So you can really scale up to sequence thousands of these types of cells.”
The trick for McGowan is to work out how to remove the noise that comes with amplifying the DNA of single cells, and make sure that the genomes that are assembled are of the highest possible quality.
“We have to clean up the genomes, because sometimes they might have bacteria present, or they might have other eukaryotes that they’ve eaten,” he explains.
“We also have to detangle the multiple genomes that might be present. That’s more challenging to automate. We don’t want to release a genome that’s actually a mix of different genomes.”
Once that process is up and running, it will be possible to begin putting the sequenced protists into their vast evolutionary perspectives.
And, for McGowan, the best part…
“A lot of this is about collaboration and openness. Making the genomes publicly available to anybody who wants to use them. That’s really good, I think.”
This article was written by Peter Bickerton and originally published on the Earlham Institute website.